This protocol describes the simple experimental procedure for Northern blot or ISH. You may need to modify this protocol if you use different reagents or instruments.
1.Introduction
The Northern blot method is used to detect specific RNAs that have been separated by size and immobilized onto a membrane (Fig 1). This method can provide specific information regarding the size of a sncRNA and possible precursor structures. Thus, it represents a valuable tool in the discovery and validation of new sncRNAs. In situ hybridization (ISH) is a technology that allows detection of specific nucleic acid sequences in tissue samples at the cellular level (Nielsen, 2012; Urbanek et al., 2015). For detection of individual sncRNA, the ISH technology determines the cellular origin of expression and provides information on expression levels in different tissue compartments and cell populations. This histological expression analysis is of crucial importance for elucidating roles particularly of sncRNAs in molecular and biological processes.
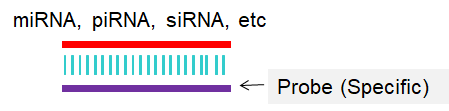
Fig 1. Scheme for Northern blot or In situ hybridization (ISH).
2. Material and Methods
2.1 Reagents and Equipments
(1) Oligonucleotide Primers. sncRNA specific primers can be designed or retrieved from sRNAPrimerDB (http://www.srnaprimerdb.com.). These primers are ordered from the company (miRNAPrimer: http://www.biootools.net/). All the primers are desalted and both UV absorbance and capillary electrophoresis are used to assess the quality of primer synthesis.
(2) Total RNA, or small RNA-enriched RNA, miRNA, siRNA, etc.
(3) Optical tube and cap strips.
(4) Digoxigenin (DIG)-labeled RNA probe in situ hybridization.
2.2 Procedure
Northern blotting involves the following steps:
(1) Total cellular RNA or mRNA is size-separated by denaturing agarose gel electrophoresis.
(2) The separated RNA is transferred onto a nylon membrane.
(3) The RNA is then detected with isotopic or non-isotopic labeled complementary DNA or RNA probe.
In situ hybridization involves the following steps:
(1) Permeabilization of cells with proteinase K to open cell membranes (around 25 minutes, not needed for tissue sections or some early-stage embryos).
(2) Binding of sncRNAs to marked RNA probe (usually overnight)
(3) Antibody-phosphatase binding to RNA-probe (some hours)
(4) Staining of antibody (e.g., with alkaline phosphatase)
2.3 Primers design
The used example sequences of RNA and DNA oligonucleotides (5’ to 3’) are listed as follows:
Experimental method I: Northern blot or ISH sRNA ID sRNA Sequence (5'-->3') Length (bp) GC (%) hsa-test-1 UGAGGUAGUAGGUUGUAUAGUU 22 36.36 PCR primer pairs Primer ID Sequence (5'-->3') Length (bp) GC (%) Tm (°C) Probe(specific) AACTATACAACCTACTACCTCA 22 36.36 53.69 Total time used: 17.250209ms.
References:
1.Northern blot analysis for microRNA (Narry Kim’s lab) :http://www.narrykim.org/legacy/Northern_blot_analysis_for_microRNA.pdf
2. Nielsen BS. MicroRNA in situ hybridization. Methods Mol Biol. 2012, 822:67-84. doi: 10.1007/978-1-61779-427-8_5.
3. Urbanek MO, Nawrocka AU, Krzyzosiak WJ. Small RNA Detection by in Situ Hybridization Methods. Int J Mol Sci. 2015, 16:13259-86. doi: 10.3390/ijms160613259.
4. Whole Mount microRNA ISH Protocol (Full Version): http://geisha.arizona.edu/geisha/documents/Whole%20Mount%20microRNA%20ISH%20Protocol%20Detailed.pdf.