This protocol describes the detailed experimental procedure for linear S-poly(A) tailed RT-PCR using SYBR Green I. The procedure begins with reverse transcription of poly(A) tailed total RNA, or small RNA-enriched RNA, miRNA, siRNA, etc. The cDNA is then used as template for real-time PCR with gene specific primers. You may need to modify this protocol if you use different reagents or instruments for real-time PCR.
1.Introduction
In linear S-poly(A) tailed RT-PCR method, formerly known as S-Poly(T) method (Kang et al., 2012) or S-Poly(T) Plus real-time PCR (Niu et al., 2015), total RNAs, including miRNAs, piRNA, or siRNA, etc. are extended by a poly(A) tailing reaction using poly(A) Polymerase and ATP. The sncRNA with a poly(A) tail is converted into cDNA through reverse transcription primed by a specific linear S-poly(T) RT primer, and then PCR-amplified using a specific forward primer and a universal reverse primer (Fig 1). The RT-PCR amplification can be monitored by real-time detection or by end-point detection for quantifying the sncRNA transcript level. The PCR amplicons can be sequenced for validating the expression of the specific sncRNA.
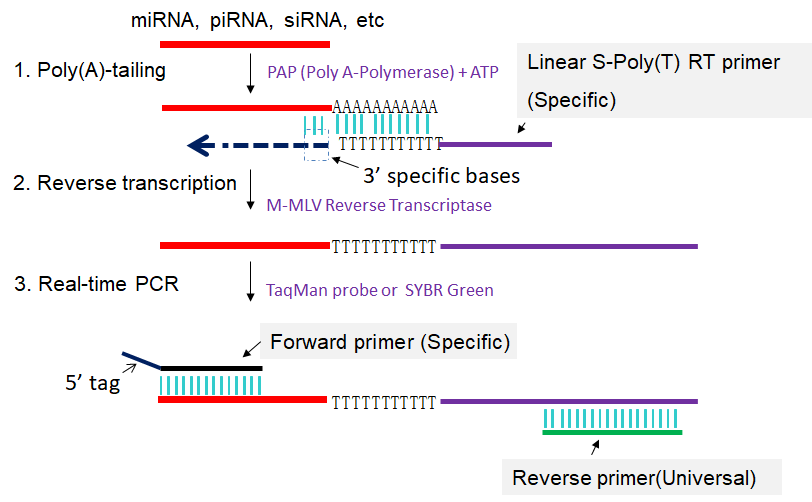
Fig 1. Scheme for linear S-poly(A) tailed RT-PCR. 1: Poly(A) tailing of sncRNA using poly(A) Polymerase and ATP. 2: Reverse transcription of poly(A) tailed sncRNA primed by specific linear S-poly(T) RT primers. 3: First-strand cDNAs of poly(A) tailed sncRNAs are amplified in PCR by using specific forward primer and reverse primer complementary to linear S-poly(T) RT primers.
2. Material and Methods
2.1 Reagents and Equipments
(1) Oligonucleotide Primers. sncRNA specific primers can be designed or retrieved from sRNAPrimerDB (http://www.srnaprimerdb.com.). These primers are ordered from the company (miRNAPrimer: http://www.biootools.net/). All the primers are desalted and both UV absorbance and capillary electrophoresis are used to assess the quality of primer synthesis.
(2) Total RNA, or small RNA-enriched RNA, miRNA, siRNA, etc.
(3) Optical tube and cap strips.
(4) Poly(A) Polymerase.
(5) M-MLV Reverse Transcriptase (RNase H-).
(6) SYBR Green PCR master mix.
(7) 50 bp DNA ladder.
(8) Quantitative PCR instrument.
(9) 3% agarose gel.
(10) Agarose gel electrophoresis apparatus.
2.2 Procedure
Reverse Transcription & cDNA Synthesis
Reverse Transcription is carried out with the Poly(A) Polymerase (PAP) and M-MLV Reverse Transcriptase (RNase H-) for RT-PCR. The following procedure is based on manufacturer’s protocol.
1. Prepare the following RNA/primer mixture in each tube. For each reaction:
| Total RNA | 0.5-1.0 μg |
| RT primer (1 μm earch) | 2 μL |
| DEPC H2O | to 3.5 μL |
2. Incubate the samples at 65 ℃ for 5 min and then on ice for at least 1 min.
3. Prepare reaction master mixture. For each reaction:
| 5 X RT buffer | 4 μL |
| 10 X PAP Buffer | 2 μL |
| dNTP (10 mmol each) | 4 μL |
| Rnase inhibitor (40 U/μL) | 0.8 μL |
| RTase (200 U/μL) | 1.2 μL |
| MgCl2 (25 mM) | 5 μL |
| ATP(100 mM) | 0.5 μL |
| PAP (2 U/μL) | 1 μL |
4. Add the reaction mixture to the RNA/primer mixture, mix briefly, and then place at room temperature for 2 min.
5. Incubate the tubes at 37 ℃ for 30 min in a PCR machine, heat inactivate at 95 ℃ for 5 min, and then chill on ice.
6. Store the 10- or 20- fold dilution 1st strand cDNA at -20 ℃ until use for real-time PCR.
Real-time PCR
The quantitative PCR reaction steps are as follows:
1. Normalize the primer concentrations and mix sncRNA-specific forward and reverse primer pair. Each primer (forward or reverse) concentration in the mixture is 5 pmol/μL.
2. Set up the experiment and the following PCR program on BioRad CFX96 Real-Time PCR. Do not click on the dissociation protocol if you want to check the PCR result by agarose gel. Save a copy of the setup file and delete all PCR cycles (used for later dissociation curve analysis).
| Stage 1: | 94 ℃, 3 min, 1 cycle |
| Stage 2: | 95 ℃, 15 s; 60 ℃ 30 s, 40 cycle |
| Stage 3: | Dissociation analysis |
3. A real-time PCR reaction mixture can be either 50 μL or 25 μL. Prepare the following mixture in each optical tube.
| 25 μL SYBR Green Mix (2X) |
| 0.5 μL cDNA |
| 2 μL primer pair mix (5 pmol/ μL each primer) |
| 22.5 μL H2O |
or
| 12.5 μL SYBR Green Mix (2X) |
| 0.2 μL cDNA |
| 1 μL primer pair mix (5 pmol/μL each primer) |
| 11.3 μL H2O |
4. After PCR is finished, remove the tubes from the machine. The PCR specificity is examined by 3.5% agarose gel using 10 μL from each reaction.
5. Analyze the real-time PCR result. Check to see if there is any bimodal dissociation curve or abnormal amplification plot.
End-point PCR
1. For end-point PCR, it is critical to amplify for an appropriate number of cycles so that 1) the PCR amplification product is readily visible on an agarose gel, and 2) the reaction remains in the exponential phase of amplification. The number of cycles required to meet these two criteria must be empirically determined. As a starting point, we recommend testing 15-25 cycles.
2. Analyze reaction products (10 μL) by electrophoresis on a 3.5% high resolution agarose gel in 1X TAE stained with dye for detecting dsDNA. Gene specific sncRNA amplicons should form discrete ~75 bp bands that are easily distinguished from smaller primer-dimer bands that may be seen in the no-template control reaction.
3. End-point PCR can be used for qualitative determination of differences in the expression of a given sncRNA between two (or more) RNA samples. In addition, a “standard curve” can be constructed to define the approximate magnitude of sncRNA expression differences between samples.
2.3 Primers design
The used example sequences of RNA and DNA oligonucleotides (5’ to 3’) are listed as follows:
Experimental method B: Linear S-poly(A) tailed RT-PCR
sRNA ID sRNA Sequence (5'-->3') Length (bp) GC (%)
Let-7a UGAGGUAGUAGGUUGUAUAGUU 22 36.36
RTprimer(specific) CAGTGCAGGGTCCGAGGTCAGAGCCACCTGGGCAATTTTTTTTTTTAACTAT 52 46.15
Fp-1 AAGCGACCTGAGGTAGTAGGTT 22 50.00 61.16
Fp-2 AAGCGCCTTGAGGTAGTAGGTT 22 50.00 62.01
Fp-3 AATCGGCGTGAGGTAGTAGGTT 22 50.00 61.80
Fp-4 ACGAGCACTGAGGTAGTAGGTT 22 50.00 61.15
Fp-5 ACGCCGTGAGGTAGTAGGTT 20 55.00 61.20
Primer Pair 1
Fp-1 5'-AAGCGACCTGAGGTAGTAGGTT-3'
||||||||||||||||||||||
5'-AAGCGACCTGAGGTAGTAGGTTGTATAGTTAAAAAAAAAAATTGCCCAGGTGGCTCTGACCTCGGACCCTGCACTG-3' (76 bp)
--------**********************
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
||||||||||||||||||
3'-ACCTCGGACCCTGCACTG-5' Rp
Possible hairpin: -1.4 kcal/mol (Gibbs free energy)
AAGCGACCTGAGGTAGTAGGTT
.....((((........)))).
Possible dimer: -5.4 kcal/mol (Gibbs free energy)
--AAGCGAC-CTGAGGTA
| |||
TGGAGCCTGGGACGTGAC
Primer Pair 2
Fp-2 5'-AAGCGCCTTGAGGTAGTAGGTT-3'
||||||||||||||||||||||
5'-AAGCGCCTTGAGGTAGTAGGTTGTATAGTTAAAAAAAAAAATTGCCCAGGTGGCTCTGACCTCGGACCCTGCACTG-3' (76 bp)
--------**********************
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
||||||||||||||||||
3'-ACCTCGGACCCTGCACTG-5' Rp
Possible hairpin: 0 kcal/mol (Gibbs free energy)
AAGCGCCTTGAGGTAGTAGGTT
......................
Possible dimer: -4.1 kcal/mol (Gibbs free energy)
----------AAGCGCCT
||||
TGGAGCCTGGGACGTGAC
Primer Pair 3
Fp-3 5'-AATCGGCGTGAGGTAGTAGGTT-3'
||||||||||||||||||||||
5'-AATCGGCGTGAGGTAGTAGGTTGTATAGTTAAAAAAAAAAATTGCCCAGGTGGCTCTGACCTCGGACCCTGCACTG-3' (76 bp)
--------**********************
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
||||||||||||||||||
3'-ACCTCGGACCCTGCACTG-5' Rp
Possible hairpin: 0 kcal/mol (Gibbs free energy)
AATCGGCGTGAGGTAGTAGGTT
......................
Possible dimer: -5.6 kcal/mol (Gibbs free energy)
-AATCGGCGTGAGGTAGT
||||
TGGAGCCTGGGACGTGAC
Primer Pair 4
Fp-4 5'-ACGAGCACTGAGGTAGTAGGTT-3'
||||||||||||||||||||||
5'-ACGAGCACTGAGGTAGTAGGTTGTATAGTTAAAAAAAAAAATTGCCCAGGTGGCTCTGACCTCGGACCCTGCACTG-3' (76 bp)
--------**********************
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
||||||||||||||||||
3'-ACCTCGGACCCTGCACTG-5' Rp
Possible hairpin: -0.1 kcal/mol (Gibbs free energy)
ACGAGCACTGAGGTAGTAGGTT
......(((.....))).....
Possible dimer: -8.3 kcal/mol (Gibbs free energy)
--------ACGAGCACTG
||||||
TGGAGCCTGGGACGTGAC
Primer Pair 5
Fp-5 5'-ACGCCGTGAGGTAGTAGGTT-3'
||||||||||||||||||||
5'-ACGCCGTGAGGTAGTAGGTTGTATAGTTAAAAAAAAAAATTGCCCAGGTGGCTCTGACCTCGGACCCTGCACTG-3' (74 bp)
------**********************
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
||||||||||||||||||
3'-ACCTCGGACCCTGCACTG-5' Rp
Possible hairpin: -3.7 kcal/mol (Gibbs free energy)
ACGCCGTGAGGTAGTAGGTT
...((....)).........
Possible dimer: -4.3 kcal/mol (Gibbs free energy)
-----ACGCCGTGAGGTA
|| ||
TGGAGCCTGGGACGTGAC
Total time used: 8.885893031s.
References:
1. Kang K, Zhang X, Liu H, Wang Z, Zhong J, Huang Z, Peng X, Zeng Y, Wang Y, Yang Y, Luo J, Gou D. A novel real-time PCR assay of microRNAs using S-Poly(T), a specific oligo(dT) reverse transcription primer with excellent sensitivity and specificity. PLoS One. 2012, 7:e48536. doi: 10.1371/journal.pone.0048536.
2. Niu Y, Zhang L, Qiu H, Wu Y, Wang Z, Zai Y, Liu L, Qu J, Kang K, Gou D. An improved method for detecting circulating microRNAs with S-Poly(T) Plus real-time PCR. Sci Rep. 2015, 5:15100. doi: 10.1038/srep15100.