This protocol describes the detailed experimental procedure for one-step real time RT-PCR using SYBR Green I. In the one-step approach, the entire reaction from cDNA synthesis to PCR amplification occurs in a single tube. You may need to modify this protocol if you use different reagents or instruments for real-time PCR.
1.Introduction
The principle of the one-step real time RT-PCR is illustrated in Fig. 1. The whole detection procedure requires four primers: RP1, RP2, P1 and P2. RP1 contains the P1 and a 11-base sequence that is complimentary to the 3′-end of target sncRNA. RP2 contains P2 and a 11-base sequence that is same with the 5′-end of target sncRNA. The total thermocycling program includes three stages. Stage 1 is the reverse transcription process. In this stage, the RP1 hybridizes with target sncRNA, and then is extended in the presence of Reverse Transcriptase M-MLV (RNase H-) and dNTPs. Since the melting temperature (Tm) of an 11-base sequence is near 37 ℃, this stage is conducted at 37 ℃. In Stage 2, after denaturing at 95 ℃, the RP2 hybridizes with the cDNA of miRNA at 37 ℃, and both sequences are extended in the presence of hot-start Taq polymerase (HS Taq) and dNTPs at 60 ℃. Stage 3 is a conventional PCR process with the primers P1 and P2. The amplification of cDNA is monitored in the real time PCR system using SYBR green I (Yan et al., 2013).
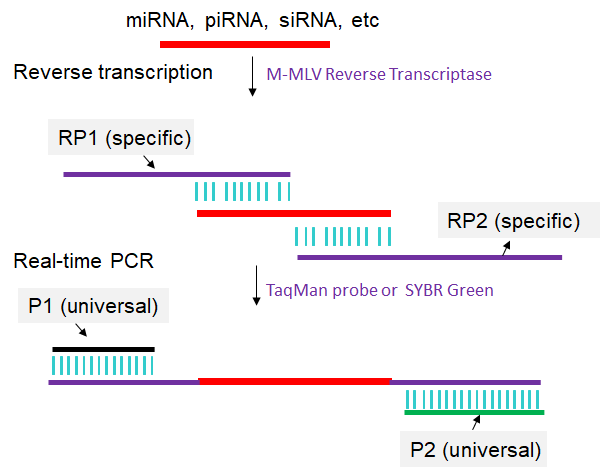
Fig 1. Scheme for one-step real time RT-PCR. For detailed experimental steps, please refer to the articles published by Yan et al., 2013.
2. Material and Methods
2.1 Reagents and Equipments
(1) Oligonucleotide Primers. sncRNA specific primers can be designed or retrieved from sRNAPrimerDB (http://www.srnaprimerdb.com.). These primers are ordered from the company (miRNAPrimer: http://www.biootools.net/). All the primers are desalted and both UV absorbance and capillary electrophoresis are used to assess the quality of primer synthesis.
(2) Total RNA, or small RNA-enriched RNA, miRNA, siRNA, etc.
(3) Optical tube and cap strips.
(4) M-MLV Reverse Transcriptase (RNase H-).
(5) SYBR Green PCR master mix.
(6) 50 bp DNA ladder.
(7) Quantitative PCR instrument.
(8) 3% agarose gel.
(9) Agarose gel electrophoresis apparatus.
2.2 Procedure
1. Prepare the following RNA/primer mixture in each tube. For each reaction:
| Total RNA | 0.5-1.0 μg |
| 5 X M-MLV Buffer* | 1 μL |
| 10 X PCR Buffer** | 1 μL |
| primer RP1 (2 μM) | 0.1 μL |
| primer RP2 (2 μM) | 0.1 μL |
| primer P1 (10 μM) | 0.1 μL |
| primer P2 (10 μM) | 0.1 μL |
| 20X SYBR Green I | 0.2 μL |
| Reverse Transcriptase M-MLV | 20 U |
| RNase inhibitor | 4 U |
| DEPC H2O | to 10 μU |
**10 X PCR Buffer (100 mM Tris-HCl, pH 8.3, 500 mM KCl, 15 mM MgCl2)
2. The real- time RT-PCR assay was conducted under the following conditions:
| Stage 1: | 45 ℃, 3 min; 37 ℃, 5 min, 1 cycle |
| Stage 2: | 95 ℃, 30 s; 37 ℃, 30s; 60 ℃ 30 s, 1 cycle |
| Stage 3: | 95 ℃, 30 s; 72 ℃, 30s, 40 cycles |
And the real-time fluorescence intensity was monitored at each cycle of the third stage. StepOne Real-Time PCR System (Applied Biosystems, USA) was used to perform the reaction.
2.3 Primers design
The used example sequences of RNA and DNA oligonucleotides (5’ to 3’) are listed as follows:
Experimental method G: One-step real time RT-PCR sRNA ID sRNA Sequence (5'-->3') Length (bp) GC (%) hsa-test-1 UGAGGUAGUAGGUUGUAUAGUU 22 36.36 PCR primer pairs Primer ID Sequence (5'-->3') Length (bp) GC (%) Tm (°C) RP1(specific) GGACGGTAGCAAGCAAAGAGTGTGAACTATACAAC 35 45.71 69.04 RP2(specific) GGGATTCTGGAAGATGATGATGACTGAGGTAGTAG 35 45.71 67.42 P1(universal) GGACGGTAGCAAGCAAAGAGTGTG 24 54.17 64.71 P1(universal) GGGATTCTGGAAGATGATGATGAC 24 45.83 59.59 Total time used: 66.628177ms.
References:
1. Yan J, Zhang N, Qi C, Liu X, Shangguan D. One-step real time RT-PCR for detection of microRNAs. Talanta. 2013, 15;110:190-5. doi: 10.1016/j.talanta.2013.02.028.